Welcome to
my page
Quentin Andres
Second Year PhD Candidate in Fisheries Science and Technology (International Program)
Kasetsart University (KU) 🇹🇭
Asian Institute of Technology (AIT) 🌏
University Côte d'Azur (UCA) 🇫🇷
About me
Introduction
👋 Hello! I’m Quentin Andres 👨💻, a second-year PhD candidate in Fisheries Science and Technology at Kasetsart University.
👉 My research focuses on the intersection of aquatic animal health, molecular biology, and genomics. Specifically, I aim to develop sustainable and low-cost vaccines 💉 for aquaculture, with a current focus on Streptococcus iniae 🦠 affecting Asian seabass 🐟. The methodologies I employ have potential applicability to a wider range of aquatic vertebrates and bacterial pathogens.
🎓 Educational Background
- Doctoral Program: Fisheries Science and Technology, Kasetsart University (Ongoing)
- MSc: Aquaculture and Aquatic Resources Management, Asian Institute of Technology
- BSc: Life Sciences with a major in Molecular and Genetic Biology (BMG), University Côte d’Azur
- Baccalauréat: Major in Life Sciences and Physics, Lycée Jacques Audiberti
🔬 Research Focus
- Development of Recombinant and Plasmid DNA Vaccines for Streptococcus iniae
- Utilization of Quality by Design (QbD) to Streamline Aquaculture Vaccine Manufacturing
- Multifaceted Approaches in Bacterial Epidemiology, Pan-genomics, and Reverse Vaccinology
📚 Current Research
My thesis is organized into multiple facets, including:
- 🧬 Low-cost vaccine manufacturing for aquaculture (recombinant platform)
- 🧬 Low-cost vaccine manufacturing for aquaculture (pDNA platform)
- 💻 Comparative genomics of Streptococcus iniae
- 🧪 Reverse vaccinology coupled with bacterial pan-genomics
- 🌡 Vaccine design, purification, and efficacy evaluation
🔎 The initial phase of my research was conducted under the Laboratory of Aquatic Animal Health Management and the Center of Excellence in Aquatic Animal Health Management, both housed within the Faculty of Fisheries at Kasetsart University.
📝 My first research article,“Comparative Genomics and Reverse Vaccinology of Streptococcus iniae: Blueprints for Affordable Aquaculture Vaccines,” introduces a novel framework for more affordable vaccines by integrating epidemiology, pan-genomics, and reverse vaccinology, coupled with Quality By Design (QbD) principles and will soon be published..
🎯 Research and Academic Interests
- Aquatic Animal Health
- Bacterial and Microbial Genomics
- Sustainable Vaccine Development
- Bioinformatics and Data Science
- Green Bioprocessing Technologies
🤝 Let’s connect
I’m committed to advancing the fields of aquaculture genomics and aquatic health management with focus on Asian aquaculture 🌏🎣.
📝 Recent Student Research and Practical Experiences
Whole Genome Assembly
- New Generation Sequencing: Performed whole genome assembly of Streptococcus iniae isolates from diseased-farmed Asian seabass fish.
- Technologies: Illumina PE short-reads and long sequencing reads (PacBio or Nanopore).
- Methodologies: Single reference mapping, de-novo assembly, reference-guided de-novo assembly, multiple reference mapping directly onto pangenome graphs.
- Submissions to public health databases: Submitted my results to NCBI under the bioproject number PRJNA933632 (Currently private, five Biosamples with short sequencing reads and GenBank assemblies will soon be released..).
Aquatic Animal Vaccines
Inactivated vaccines: Developed and evaluted oral-oil-encapsulated and immersion monovalent and bivalent vaccines for Streptococcus agalactiae and Aeromonas veronii for Nile tilapia. Found that for those formulations specifically, immersion was more effective in conferring protective efficacy for a few months and that oral vaccines were more appropriate to be given as a booster after primer vaccination.
mRNA vaccines: Explored mRNA vaccine design and manufacturing against TiLV for Nile tilapia (Project stalled)
📝 Older Student Research and Practical Experiences
Production of Biomolecules
- Research Objectives: Compared eukaryotic and prokaryotic vectors for recombinant GFP and B-galactosidase expression.
- Cellular Models: HEK 293 immortalized cells vs Escherichia coli K-12 strain with T7RNAP+ capabilities).
- Specific Activity, Qualitative and Quantitative Measurements: Employed fluorescence microscopy and plate reader for fluorescence measurements, BSA for protein quantification, SDS-PAGE for protein quality, and indirect enzymatic assays using HRP for color development.
Cellular Signaling and Apoptosis
- Research Objective: Explored apoptosis mechanisms and cellular signaling pathways, as part of a team.
- Cellular Models: Utilized the Drosophila S2R+ cell line.
- Cell Growth & Survival: Monitored using cell counting to ensure adaptability to laboratory conditions.
- Transfection Efficacy: Evaluated using Effectene transfection reagent on S2R+ cells, incorporating both an empty vector and a GFP-containing expression vector.
- Induction of Apoptosis and Necrosis: Facilitated by the addition of staurosporine to the cell culture medium; optimized the concentration range for apoptosis induction.
- Imaging Techniques: Utilized confocal microscopy with propidium iodide tagging in S2R+ cells to study apoptosis signaling pathways-mitochondrial pathway post-staurosporine treatment.
Cardiovascular physiology
- Research Objective: Investigated the regulatory mechanisms of blood vessel diameter, as part of a team.
- Model Used: Isolated ring of aorta from a rat.
- Agents Administered: KCl, verapamil, Isoprotérénol, phenylephrine, various concentrations of NA (noradrénaline), and prazosin.
- Key Insights: Recorded variations in vessel diameter in response to administration of agents affecting calcium ion channels and hormonal activity.
Plant Biology
- plant physiology: Worked on stomatal opening in plants and the role of Abscisic Acid (ABA) and Gibbrellic acid (GA).
Bioinformatics and Computational Biology
- Venom Gland mRNA Discovery: Computational approach in discovering and analyzing a new mRNA in the venom gland of the endoparasitoid insect Aphidius colemani, as part of a team.
- Homology Hypothesis: Utilization of bioinformatics tools for sequence alignment and homology search.
- ORF Analysis: Use of EMBOSS tools to search for possible Open Reading Frames.
- Functional Analysis: Database use for developing functional hypotheses for proteins.
- Motif and Domain Identification: Employed ScanProsite and InterProScan for motif and domain identification.
🛠 Methodologies and Tools
👨💻 Programming Languages
- Proficient in data manipulation, analytics, github for control version, and bioinformatics scripting using Bash, R, and Python. Some experience in Perl.
Experience Levels
🟩🟩🟩🟩🟩 R
🟩🟩🟩🟩🟨 Bash-command line
🟩🟩🟩🟨🟨 Python
🟩🟩🟨🟨🟨 Perl
🐧 Operating System
- Mostly using Linux for applied bioinformatics, development and data analysis.
🔬 Molecular Techniques and Microbiology
- Molecular Biology Skilled in various cloning techniques such as bacterial transformation, IVA Cloning, and Golden Gate Assembly. Experienced in DNA Gel-Electrophoresis for molecular separation.
- Proteomics: Western Blotting (WB), SDS-PAGE, Non-reducing Capillary Gel Electrophoresis (CGE)
- Bioprocessing: Protein purification, chromatogrpahy
🧪 Fundamental Lab Skills
- Animal: Eukaryotic cell culture and viability assays (MTT) and trypan blue assay, transfections in HEK 293 and other cells.
📊 Analytics
- Immunology: Expertise in Real-Time Quantitative PCR (RT-qPCR), Enzyme-Linked Immunosorbent Assay (ELISA) and its variations. Limited experience with Fluorescence-activated cell sorting (FACS) and cytotoxic activity tests (cytotoxic activity of T cells and Natural Killer cells measured by chromium release assay (CRA)).
🐁/🐟 In-vivo Studies
- Basic toxicology: Histopathology, basics of animal physiology. Familiarity with aquatic animals mucosal and systemic immune responses.
- Basic immunology and analytics: Experience in blood count, serum preparation, and survival challenges for immunological assays and screenings in fish.
- Functional Renal Exploration: Experience with rat models to study renal functions. Techniques involved: jugular and carotid cannulation. Used various substances including inuline, mannitol, and furosemide.
📜 Regulatory and Pharmaceutical Development
- Quality by Design (QbD): Employ QbD methodologies in the development and optimization of recombinant vaccines.
- Regulatory: Familiar with guidance frameworks and regulations in particular:
🎯 I am focused on advancing the fields of aquaculture genomics and aquatic health management.
Download My Résumé. (coming soon..)
👉 Feel free to 📩 email me for collaborations or discussions.
You can also find me on 🤝 LinkedIn, 💻 Github and check out some of my publications in my 🌐 academic page below.
The following facet presents the different skills that I master or very familiar with:
Skills
In-silico cloning (DNA Cauldron)
PyDNA
Snapgene
NEB Golden Gate Tool
Benchling
PCR
In-vivo assembly cloning
Golden Gate Assembly cloning
Separation of DNA by gel-electrophoresis
Purification of nucleic acid sequences (gDNA)
Purification of nucleic acid sequences (pDNA)
Purification of nucleic acid sequences (RNA)
Recombination & mutagenesis
Transcriptional fusions
Bacterial transformations
DNA probe hybridization on nitrocellulose membrane
qRT-PCR
qPCR
Preparation of culture media for Escherichia coli
Gram-staining
Bacterial transformations
Cellular culture (animal)
MTT Assay
Trypan blue assay
Transfection
Propidium Iodide assay
Fluorescence microscopy
Confocal microscopy
Purification of proteins by affinity to GST
SDS-PAGE
BCA assay
Bradford assay
Western blot
ELISA & ELISPOT
RefSeq
UniprotKB
RegulonDB
VICTORS
VFDB
PATRIC
Immune Epitope Database (IEDB)
NCBI
EBI
GenBank
Ensembl
NCBI entrez
JSON
XML
SQL
Kraken2
SeqAdapt
SeqPurge
Trimmomatic
Awk
seqtk
FastQC
QUAST
Bandage
Unicycler
SPAdes
Mauve
Mummer
Bowtie2
Picard
Bamtools
bcftools
PILON
Velvet
Snippy
BWA-MEM
fastANI
QUAST
Mash
Integrated Genome Viewer (IGV)
PopPUNK
fastbaps
GrapeTree
Microreact
DBScan
KNN
K-means clustering
Prokka
PGAP
CRISPRTyper
ISmapper
PanISa
BEDTools
BoBro2.0
GOST
KEGG
KRONA
Progressive Mauve
Progressive Cactus
PhyML
IQ-Tree 2
ggtree
Ape
Phytools
Castor
Gubbins
ClonalFrameML
HyPhy
vg Toolkit
ODGI
PGGB
vg (Sequence Tube Maps)
Roary
Piggy
panX
Pandora
Panaroo
Cytoscape
ForceAtlas2
PPanGGOLiN
MAFFT
Clustal Omega
Diamond
BLAST
SKA2
Bowtie2
INTERPROSCAN
TIGRFAM
SIGNALIP
PRINTS
Pfam
Antifam
TMHMM
MobiDBLite
eggNOG 6.0
Circos
R Suite
Bioconductor
Github
GNU parallel
Understanding of data structures
Inference and Modeling
Data Wrangling
Exploratory Data Analysis
Linear Regression
Principal component analysis
(recombinant protein vaccines and bioprocessing)
Comprehensive analysis of up-to-date public knowledge on recombinant protein vaccines
Protein expression systems
Protein purification systems
Vaccine efficacy
Bioprocess optimization
Quality by Design (QbD)
(plasmid DNA vaccines and bioprocessing)
Comprehensive analysis of up-to-date public knowledge on plasmid DNA vaccines
Vector design
Optimization of expression
pDNA purification methods
Bioprocess scale-up
Vaccine efficacy
Quality by Design (QbD)
(pan-genomics and Firmicutes)
Comprehensive analysis of up-to-date public knowledge on pan-genomics and of Firmicutes
Comparative genomics and functional genomics
Streptococcus Mutans, Bovis, Pyogenic group
Streptococcus iniae
Streptoccocus agalactiae (GBS)
Streptococcus pyogenes (GAS)
ggplot2
ggpubr
dplyr
tidyverse
plotly
Markdown
Blogdown
Shiny
I am familiar with those topics to a certain point and have ever raised juveniles and adult Nile Tilapia during vaccine trials
Recirculating Aquaculture Systems
Aquaponics systems
Biofloc systems
Hatchery technologies
Catfish farming
Carp farming
Tilapia farming
Shrimp farming
Seafood business
Fish nutrition
Feed formulation design
Farm design
Sustainable aquaculture and SDGs
French native
English fluent
Thai (ภาษาไทย) B2
Spanish B1
Slovak A2
Trainings & MOOCs
Introduction to Linear Models and Matrix Algebra.
Statistical Inference and Modeling for High-throughput Experiments.
High-Dimensional Data Analysis
Conferences & Workshops
Featured Publications
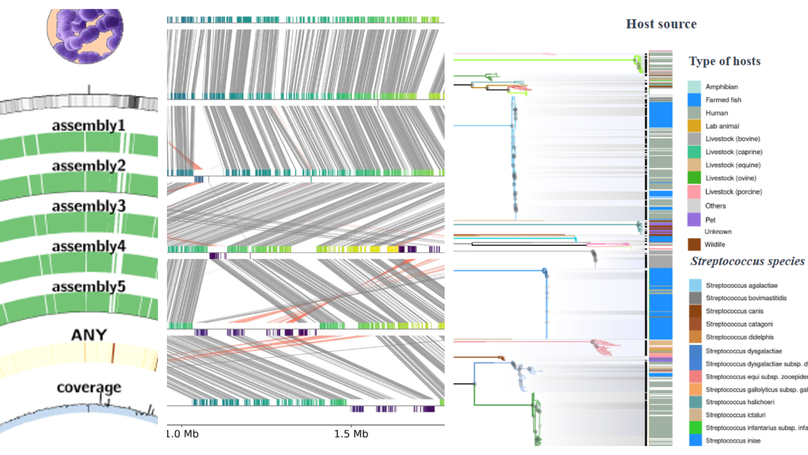
The high cost of vaccines relative to antimicrobials in aquaculture limits their adoption, particularly in low-value freshwater fish that nonetheless play a crucial role in global food security. Current vaccines, killed or live attenuated are highly effective but are no longer cost-efficient. Here, we present a new framework that leverages epidemiology, pangenomics, and reverse vaccinology with Quality By Design (QbD) principles to design more affordable vaccines. Targeting a range of aquatic streptococcal species, we emphasize the fish pathogen Streptococcus iniae. We identify a subset of protein antigens from the proteome that are well-adapted for low-cost mass production using QbD manufacturing strategies. This integrated approach is a crucial step towards making vaccines more accessible and cost-effective for global aquaculture, ultimately aiding in combating antimicrobial resistance and ensuring food security and consumer health.
Summary presentation of my PhD thesis for the part on genome sequencing and Analysis, comparative genomics of Streptococcus iniae and explaining about the manufacturing strategies of recombinant proteins vaccines (pDNA vaccines are not included..)
Aquaculture has made significant strides in providing a sustainable and efficient protein source. However, bacterial infections such as those caused by Streptococcus agalactiae and Aeromonas veronii remain a persistent challenge, affecting the health and productivity of Nile tilapia (Oreochromis niloticus) fingerlings. This study aimed to assess and compare the efficacy of two bivalent inactivated vaccines administered through different methods: oral vaccine followed by an oral booster (OR+OR) and immersion vaccine followed by an oral booster (IM+OR).
Contact
- quentin.ls.andres@gmail.com
- 66855167216
- 50 ถนน งามวงศ์วาน แขวงลาดยาว เขตจตุจักร กรุงเทพมหานคร 10900, Bangkok, Bangkok Province 12100
- I work from office and from home.
- Not fixed, Monday-Friday 10:00 to 18:00
- Access my website
- Github
- Linked-in








